For the first aim ‘Innovative Microscopy and Guidance of Cells’, we will implement, integrate and advance innovative technologies that will enable visualisation, quantification and manipulation of molecular processes in living cells in tissues at high spatial and temporal resolution.
This Technological Aim will be organised into two work packages:
TechWP1- Bridging scales: High-resolution microscopy in tissue
Here, we will focus on bridging between spatial and temporal scales and establish methodologies for high-resolution, subcellular imaging in multicellular systems.
We will achieve this by using and further enhancing the following methods:
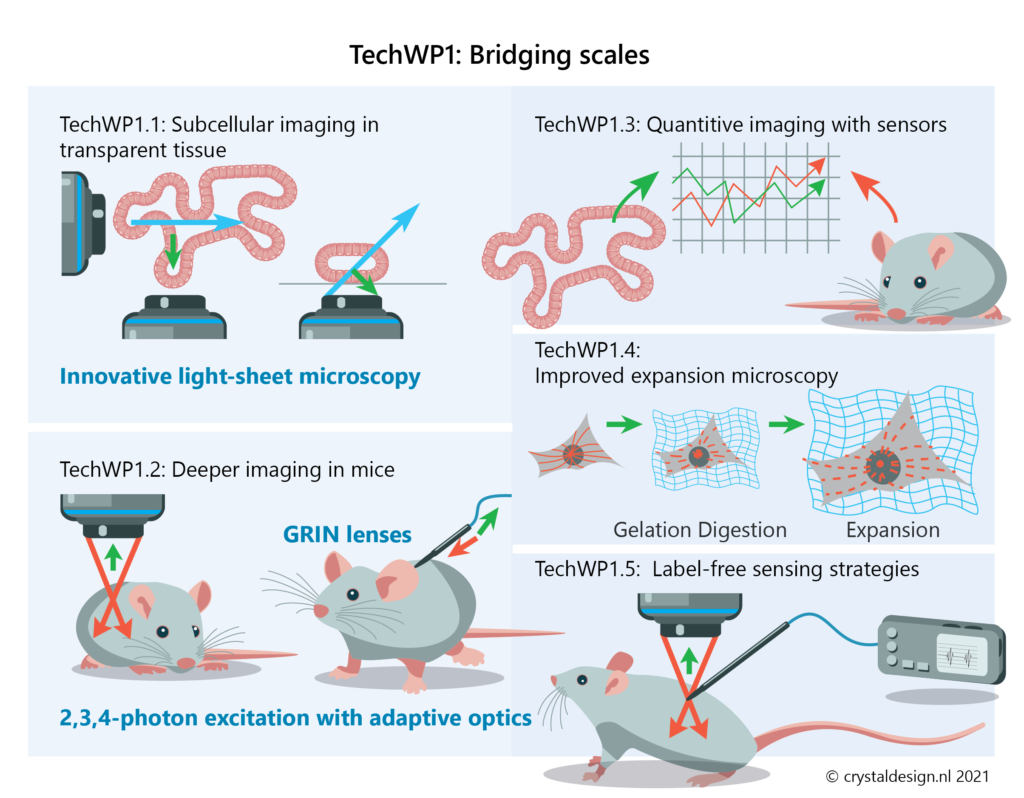
- Light-Sheet Microscopy for transparent models (TechWP1.1)
- live-tissue Multiphoton Microscopy in mice (TechWP1.2)
- quantification methods for fluorescent sensors to quantify protein activity in live cells across different model systems (TechWP1.3)
- further develop Expansion Microscopy to map the subcellular structures in tissue with nanoscopic resolution (TechWP1.4)
- establish label-free sensing strategies for drugs and biomarkers that will enable accurate concentration measurements during live-cell imaging in tissue or animals (TechWP1.5)
TechWP2 – From observing to controlling: Automated control of biological systems
Optogenetics and photopharmacology now provide powerful opportunities to monitor and modulate cellular organisation and functioning with high spatiotemporal resolution in multicellular organisms. Systematically exploring how cellular systems respond to various perturbations will reveal how cells operate inside tissues. This will also provide opportunities to remotely control cellular behaviour to steer tissue organisation and functioning.
Such experiments will allow to address our biological questions with unprecedented precision. However, the majority of available technologies have merely reached proof-of-principle level, due to the many challenges associated with applying them to in vivo systems.
In this Work Package, we will develop event-driven imaging and high-precision control strategies. These will enable real-time manipulation of key aspects of the biological systems that will be studied (smart microscopy).
Ultimately, our goal is to establish microscopy-guided automated real-time control of biological processes. This will enable automatic identification of certain cellular states or events (e.g. a cell in mitosis, a cell moving out of a niche) within a tissue. Automatic and iterative light-driven perturbations can then be used to alter the cellular state or event (e.g. altering the orientation of the mitotic spindle, returning the cell to the niche).
To achieve this we will focus on the following aspects:
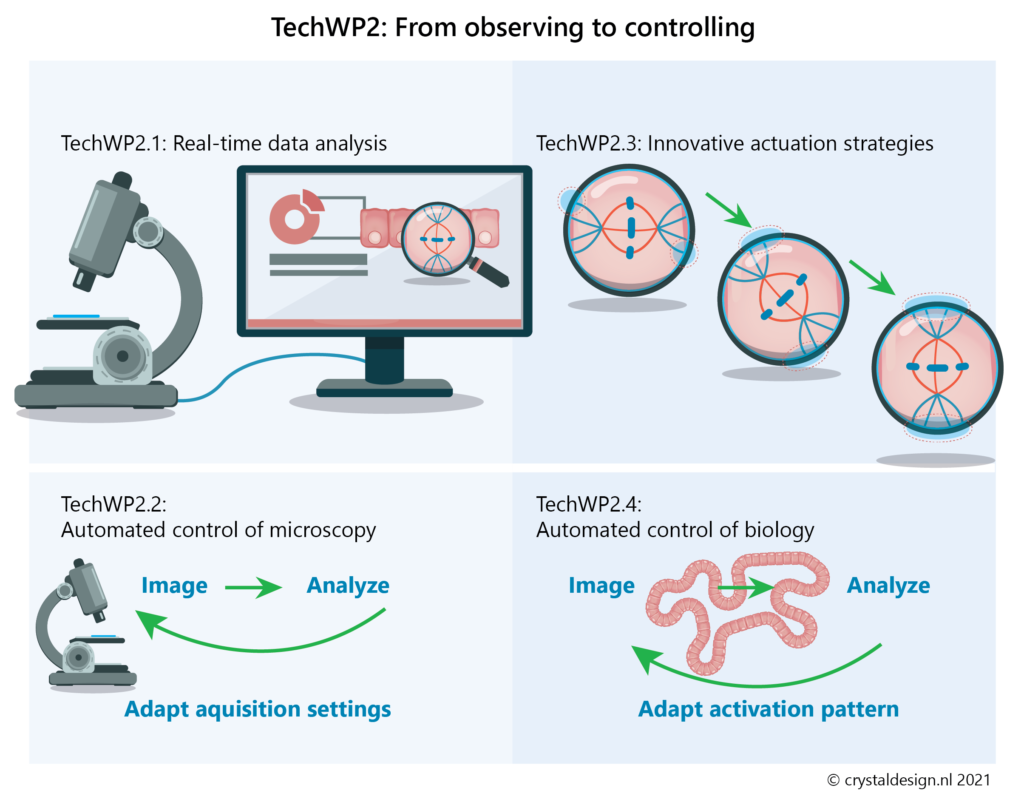
- robust procedures for real-time image analysis to identify and follow specific events during the experiment (TechWP2.1)
- optimise the acquisition settings during experiments (e.g. after a mitotic cell has been identified, and to return to more global settings after mitosis is completed; TechWP2.2)
- develop and optimise novel actuation strategies to control specific pathways or processes (TechWP2.3)
- achieving automated control of biology (TechWP2.4)
Together, this set of technologies will be extremely valuable to reveal how specific cellular events contribute to tissue formation and (mis)functioning and thus answer the biological questions in BioWP1-3.
Various already existing optogenetic strategies developed within our consortium (e.g. for organelle and spindle positioning) will be used to establish automated control strategies, while they will also be applied from the start for the more low-throughput experiments in the BioWPs.