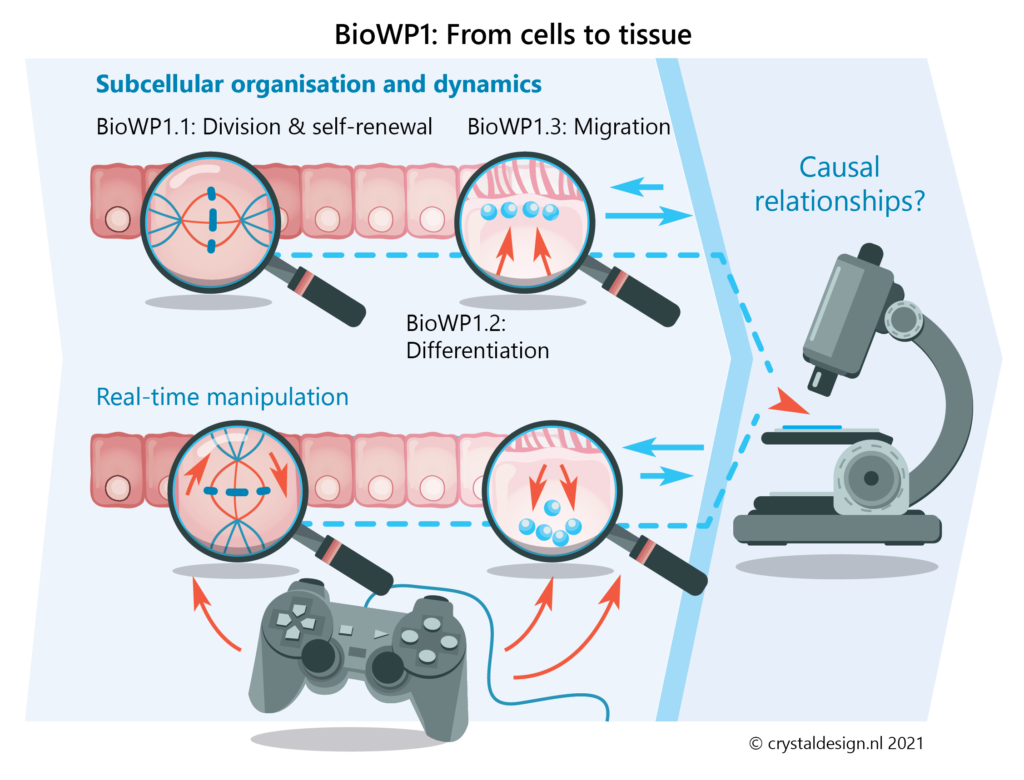
Implementation of existing technologies and further development of these will enable us to uncover the cell biology that underlies tissue self-organisation. In particular, we will focus on cell renewal, differentiation, migration, and the interplay between them.
These processes depend on changes in cytoskeletal and membrane organisation, organelle positioning and functioning, and (re)distribution and dynamics of signalling molecules. Such intracellular events, translate to intercellular changes in cell adhesion or cell-cell communication that together shape tissue organisation and function.
We will dissect these mechanisms in living cells within tissues. We will apply automated real-time (optogenetic) control strategies to steer tissue-forming processes. These efforts will be integrated with studies of pathological tissue dynamics and the impact of drugs.
This Biological Aim will be organised into three work packages:
BioWP1 – From cells to tissue: Cell biology of tissue self-organisation
Here, we will bridge scales from intravital imaging of tissues and complex tissue (organoid) cultures to high-resolution imaging of cellular constituents. This will reveal the basic self-organising principles governing tissue formation at the cellular and subcellular level.
We will focus on three key cellular behaviours:
- cell renewal
- cell differentiation
- cell migration
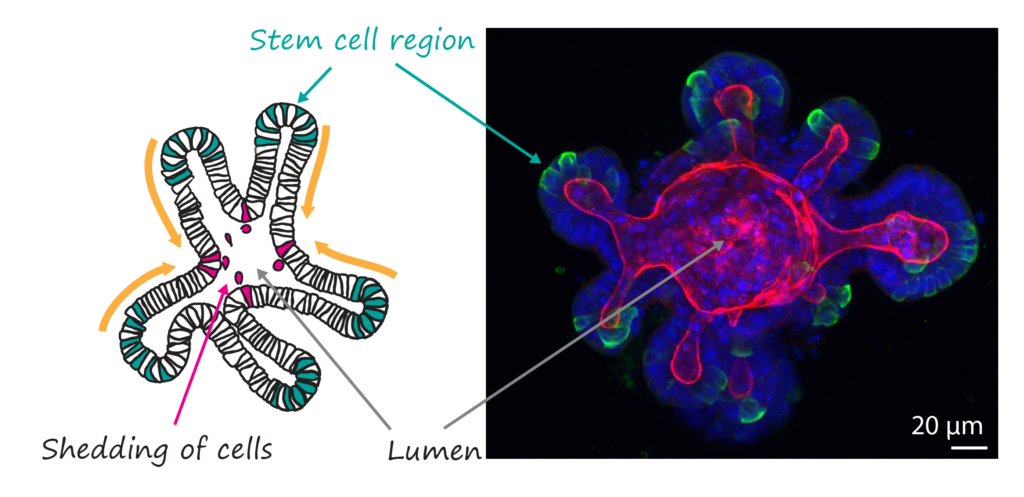
Cells in our bodies exist in complex networks. With the right stimuli, cells have the capacity to self-organise into ordered tissues with specific functions. Despite all we learnt about cells from 2D cultures, it is becoming increasingly clear that even well-studied cell types display very different architecture and dynamics within native tissues because they cannot self-organise in 2D.
Novel complex tissue culture approaches ranging from organoids to organ-on-a-chip systems now enable high-resolution live imaging of tissue self-organisation in a 3D environment. Here, we will use in vitro 3D models of intestine, breast and airways that each show striking features of self-organisation.
Using different probes and labelling techniques we will track self-organisation and identify the 3D location of individual fluorescently (Confetti) labelled cells and their progeny over time. We will map key transitions such as symmetry breaking, polarity establishment and cell repositioning.
The in vitro studies will be complemented by in vivo studies, where high spatial resolution is more challenging but where the full complexity of tissue biology is present. To achieve this, we will perform advanced microscopy using Light-Sheet, Multiphoton and Expansion Microscopy. Established, as well as newly developed microscopy workflows (such as smart microscopy developed in Aim1) will be key to the success of this aim.
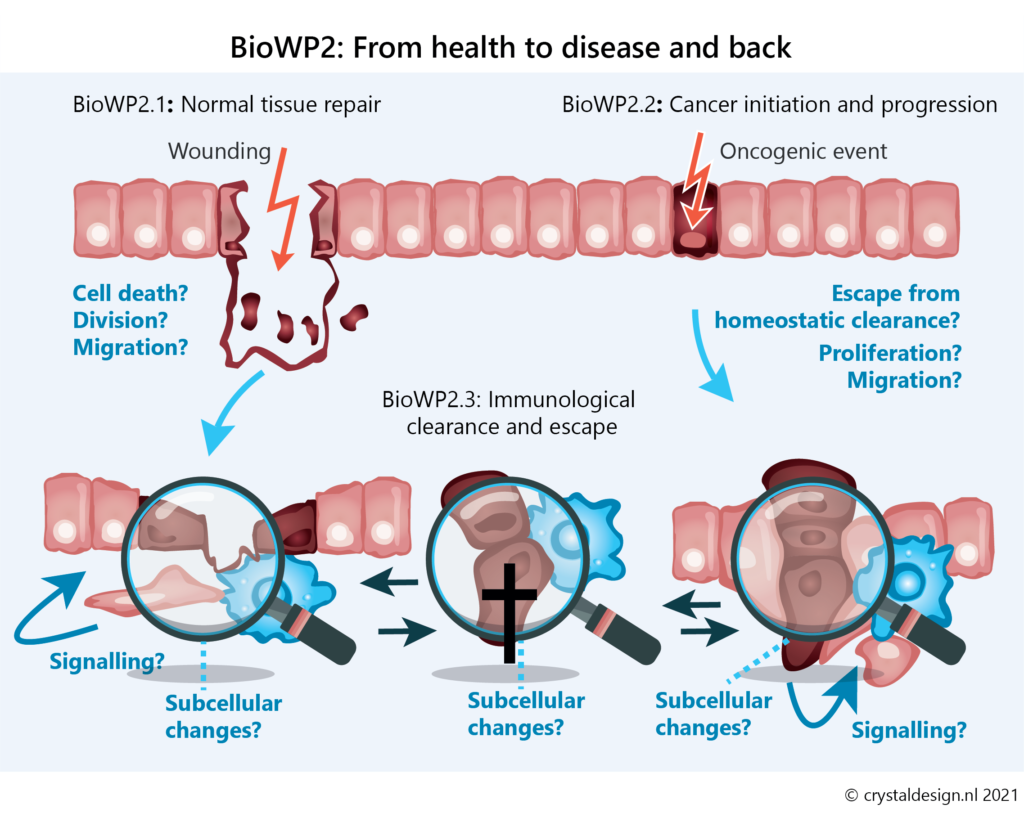
BioWP2 – From health to disease and back: Cell biology of tissue perturbation and repair
In this work package we will employ multiscale in vitro and intravital imaging to uncover how alterations in tissue self-organisation control repair at the cellular and tissue level. We will investigate how these processes are misguided to promote cancer. And we will unravel how immune cells interact with damaged and transformed epithelial cells to promote tissue repair and eliminate tumours.
The relationship between cell fate switches and dynamic alterations in subcellular architecture remains a crucial but poorly understood area of cell biology. Here, we will advance this research area by connecting recent developments in intravital imaging and in vitro models that accurately recapitulate injury-repair cycles (like intestinal organoids).
BioWP3 – Seeing drugs in action: Understanding and improving therapeutic interventions
Molecular and cellular drug targets are typically identified in biochemical assays, experiments with homogeneous cell cultures and preclinical end-point analyses. However, very little is known about how drug treatments affect the subcellular architecture and function of target and non-target cell populations within tissues in vivo.
In this work package we will:
- determine how drugs affect their intracellular targets in tissues at therapeutically relevant concentrations
- investigate how drug treatments modify the self-organising properties of cells and tissues
- determine which subcellular and cellular targets are causal to therapeutic effects of drugs and drug combinations
- elucidate how drug resistance emerges and how drug effects are modulated by the immune system
These investigations have the potential to directly impact drug efficacy. Based on our findings, co-targeting regimes may be developed to overcome drug resistance. Importantly, we will use patient-derived organoids as a tissue-mimicking platform to assess drug efficacy under clinically relevant drug concentrations. We will also validate drug efficacy, the identified mechanism(s) and toxicity using in vivo models.
Better understanding of pharmacological mechanisms within tissue is essential to improve drug success in clinical trials. Furthermore, this understanding becomes paramount for using drug combinations, repurposing, and overcoming drug resistance. Establishing the use of patient-derived organoids in drug trials will further allow for more personalised treatments.